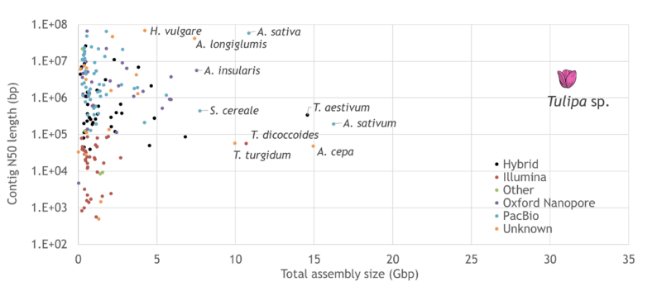
CropXR launched. NWO contributing € 15 million. New institute to develop ‘smart breeding’ method for more resilient, sustainable and climate-adaptive agriculture.
The Netherlands Organisation for Scientific Research (NWO) will contribute 15 million euros to a CropXR research programme into ‘smart breeding’ of more resilient crops. NWO’s grant marks the start of the new Dutch institute, which will integrate plant biology, computational modelling, and artificial intelligence into ‘smart breeding methods’. Those will be used to develop crop varieties that are more resilient to climate change and less dependent on chemical crop protection.
In CropXR, four Dutch universities and dozens of plant breeding, biotech and processing companies will collaborate on basic scientific research, data collection and data sharing, education, and advancing broad application of the results.
More sustainable and climate-proof agriculture
“The fact that NWO makes a substantial investment in PlantXR, a ten-year research programme central to CropXR, demonstrates its confidence in the mission of the new institute,” says Guido van den Ackerveken, Scientific Director of CropXR. “I am very proud of the collaboration of so many different partners and stakeholders in this programme. Both universities, companies and stakeholders representing the green sector feel united in contributing to our mission: making crops more resilient, sustainable, and climate-adaptive. Thanks to the NWO-contribution, we can now start our research needed to reach our common goals and make this journey a great success.”
Miriam Luizink, chair of the Programme Committee of NWO’s KIC innovation program, said: “With Long-Term Programmes, NWO offers long-term funding for strategic research by public-private consortia. It provides substantial funding, which offers a powerful boost to the development of a scientific field. In this particular case, it gives PlantXR, as part of CropXR, a better chance to successfully develop resilient crops that can remain productive even under harsh environmental conditions.”
Genetwister Technologies and its shareholder companies Bejo Zaden, Dümmen Orange, East‑West Seed, Known-You Seed Co., Ltd and Sakata Seed Corporation are all closely involved in the CropXR Institute. ”The launch of CropXR means that scientists in academic and industrial organizations will collaborate closely in multidisciplinary research programs to develop resilient crops and contribute to a sustainable society. We are looking forward to contributing our expertise regarding breeding for complex plant traits to realize these goals”, as said by Nikkie van Bers, Head of Innovation and Application of Genetwister Technologies.
Integrating plant biology, computational modelling, and AI
In the research program co-funded by NWO, academic research groups and mainly Dutch plant breeding companies will collaborate in developing a ‘smart’ method that will enable breeders to make crops more resistant more quickly. By innovatively integrating modern plant biology with artificial intelligence (AI) and computational modelling, they will learn to understand and predict how plants, using a complex interplay of multiple hereditary factors, can better withstand stress conditions. Using this knowledge, they will then develop stronger, more resilient varieties of several model crops.
At present, developing more resilient plants is very difficult and takes a very long time. The newly developed ‘smart breeding’ method is anticipated to speed up the work of both traditional plant breeders and those who apply new breeding technologies.
Infrastructure, education, social dialogue, and marketing
In addition to research, CropXR will invest in shared data infrastructure. Together with universities of applied science it will also work to promote training for professionals. It will advance broad application of its ‘smart breeding’ method by interacting and working with breeding companies and other stakeholders such as consumer organisations, environmental and development NGOs, both in the Netherlands and abroad.
The sooner a wide range of seeds, tubers, and other starting materials for more resilient crops will be available in various markets, the sooner farmers and consumers will benefit.
More resilience urgently needed
Speeding up the development of extra-resilient (XR) crops is urgently needed worldwide because many crops are faced with more extreme conditions such as heat, drought, flooding, and pathogens, that are all more extreme because of climate change. At the same time, environmental regulations are becoming stricter, which will reduce farmers’ ability to treat their crops with chemical fertilizer, pesticides and other plant protection products. For agricultural production to become sustainable in coming decades, it will need crops that are more resilient.
More information:
More information from CropXR is available on the new institute’s website: www.cropxr.org.
You can contact us at info@cropxr.org.
About the CropXR Consortium
CropXR is an initiative of four Dutch knowledge institutions (Utrecht University, Wageningen University and Research, the University of Amsterdam, and Delft University of Technology) and Plantum, the umbrella organisation of approximately 250 Dutch-based producers of plant propagation materials who together are global export market leaders in starting materials such as vegetable seeds, seed potatoes and ornamental crops.
Apart from NWO’s Long Term Programme, CropXR will also receive funding from the Dutch National Growth Fund (a funding proposal submitted by the Dutch Ministry of Agriculture, Nature and Food Quality (LNV) was approved in 2022). Financial contributions also come from the Foundation for Food & Agricultural Research (FFAR), the private consortium partner and the four universities.
CropXR will help to make agricultural production less vulnerable to climate change and less dependent on artificial fertilizers and chemical pesticides, thereby also creating growth opportunities for relevant economic sectors in the Netherlands.
In addition to knowledge institutions, dozens of public and private partners participate in CropXR’s work, including ‘green’ universities of applied sciences, biotechnology companies, processing industries and large and medium-sized plant breeding companies.
About Genetwister Technologies B.V.
Genetwister is an innovative Dutch biotechnology company founded in 1998 specialized in molecular breeding and bioinformatics of agricultural, horticultural and ornamental plants. Through our research projects we help our customers improve crop quality to enable more sustainable farming and a more reliable food supply for the future. Our shareholders are five internationally renowned vegetable and ornamental breeding companies we are proud to serve.
For more information, please contact Tina Graafmans at secretariaat@genetwister.nl.