Application of graph-based pangenomes to accelerate plant breeding
Researchers at Genetwister Technologies have generated a graph pangenome of seven cucumber varieties, capturing large-scale structural variation of this important crop. The cucumber pangenome is the culmination of multiple years of effort to bring the possibility of complete genetic characterization of crops closer to plant breeders.
The cucumber pangenome is presented by Bart Nijland at the AGBT-Ag meeting on March 27-29 in San Antonio, Texas.

Pangenomes
Conventional genetic analyses of crops rely on single references to which all other accessions are compared. However, in the past decade we have come to appreciate that structural genetic variants are ubiquitous not only in wild material but also in cultivated germplasm, and that these are often associated both with important agricultural traits and challenges in breeding programs. This significant fraction of genetic diversity is missed in the traditional single-reference analyses. However, it can be captured by characterizing germplasm with reference-quality genome assemblies and using advanced tools that allow comparison of multiple whole genome reference sequences, termed graph pangenomes. To showcase the use of pangenomes for plant breeding, researchers at Genetwister generated four new cucumber genome assemblies and integrated them in a graph pangenome with three publicly available cucumber assemblies.
“The pangenome approach enables us to investigate the genetic diversity of many accessions, in a computationally efficient way”, explained Lidija Berke, team Lead Bioinformatics and Software Development. “It is currently one of the most exciting developments in plant breeding that will speed up the development of improved varieties, harnessing the newly discovered genetic diversity.”
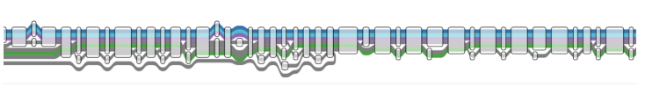
(Part of) a structural variant in one of the 7 genomes in the pangenome graph
High-quality genome assemblies
As graph-based pangenomes compare entire chromosomes, it is important that the genome assemblies incorporated into pangenomes are of high quality. To obtain high quality genome assemblies, researchers at Genetwister developed an isolation protocol for high-molecular weight DNA. The four cucumber samples were sequenced on PacBio HiFi sequencers. The high-accuracy long reads are important for resolving repeat-rich regions that are common even in small plant genomes such as cucumbers. Plant breeding companies are excited about the possibilities that pangenomes provide to accelerate plant breeding. Saulo Aflitos from Bejo Zaden comments: “For researchers, we expect pangenomes to simplify finding shared or unique regions in a panel of individuals against a panel of references, a frequent occurrence in breeding. For bioinformaticians, we expect that pangenomes reduce the (personnel and computational) costs of computation, storage and data management since all variants, all references and all queries are centralized in a single place, so we are excited about the developments”.